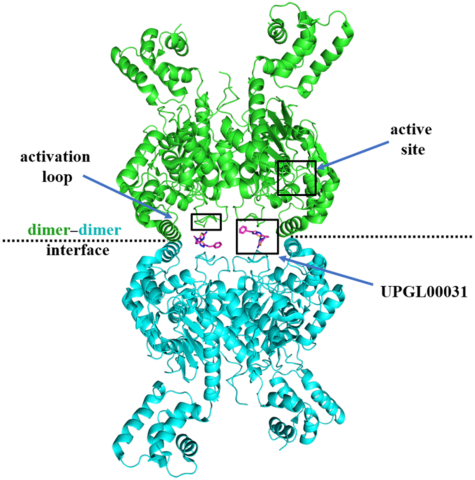
Cancer cells often overexpress glutaminase enzymes, in particular glutaminase C (GAC), which resides in the mitochondria and catalyzes the hydrolysis of glutamine to glutamate. High levels of GAC have been observed in aggressive cancers and the inhibition of its enzymatic activity has been shown to reduce their growth and survival, both in vitro and in mouse models. Numerous GAC inhibitors have been reported, with the most heavily investigated being a class of compounds derived from the small molecule BPTES (bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide). However, thus far, no GAC inhibitor has been approved for cancer treatment. Our analyses of 11 new X-ray crystal structures (Figure 1) for GAC complexed to members of the UPGL series of the BPTES compound family of varying inhibitory potency, using crystals obtained under cryo-cooled conditions, showed that the binding contacts for the different inhibitors were largely conserved. Thus, despite these extensive crystallization efforts, the molecular determinants that dictate potency for this class of GAC allosteric inhibitors were not evident.
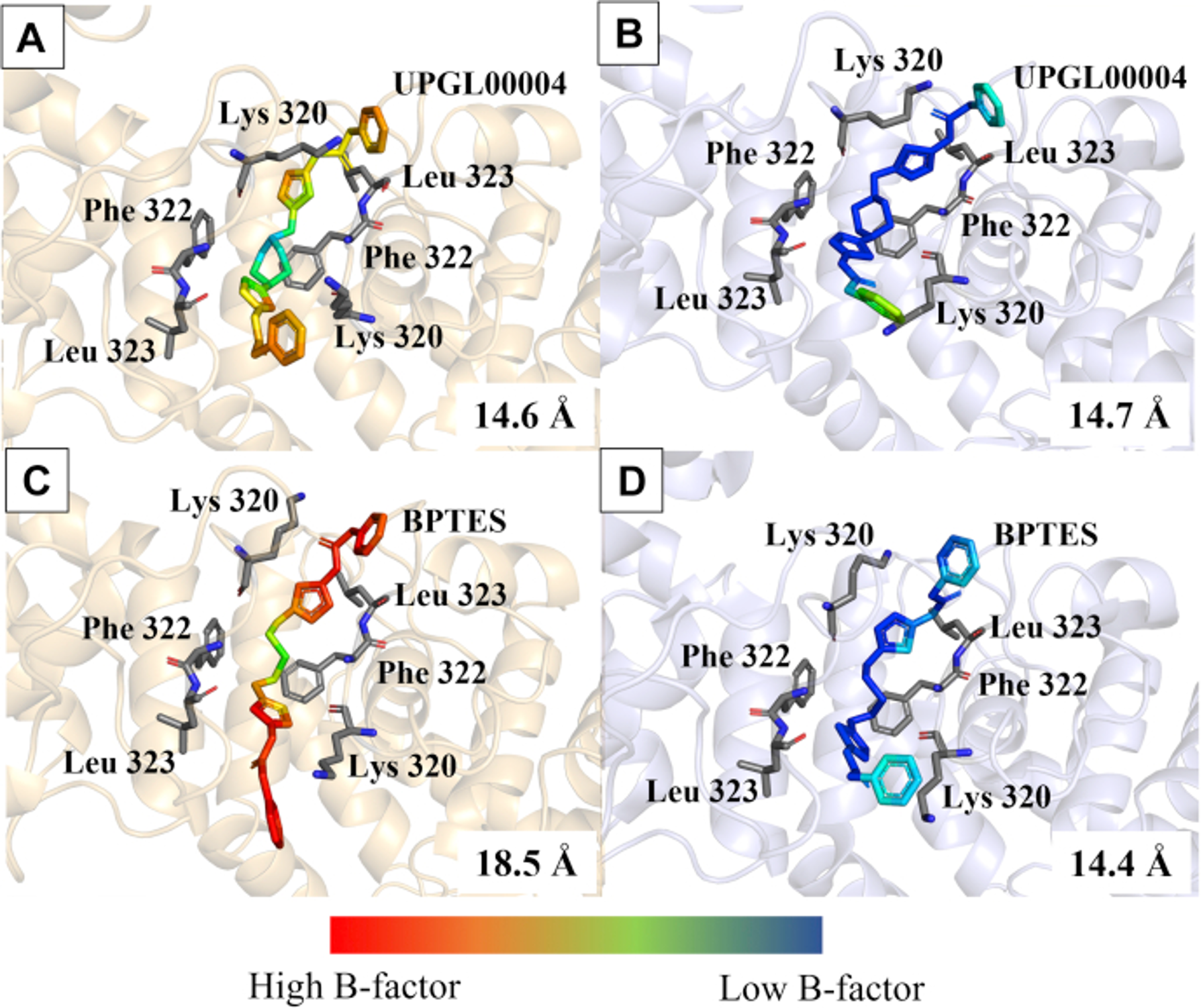
Given that the traditional cryo-cooled crystallography could not differentiate between drugs with different potencies, in our recent publication we turned to serial X-ray crystallography (SSX), which collects data from dozens of individual crystals at room temperature to achieve a high-resolution structure, and offers the potential to reveal dynamic ligand-binding states not detected when using cryogenic methods. We took advantage of this method to examine two allosteric inhibitors of GAC with different potencies, BPTES (IC50 = 371 nM) and UPGL0004 (IC50 = 29 nM). The room temperature X-ray crystal structure for GAC complexed to BPTES showed two distinct poses, with one of the BPTES molecules exhibiting an extended conformation, and the other more closely resembling a prior cup-shaped structural image of this inhibitor (Figure 2). By contrast, both molecules of UPGL00004 bound to GAC in the room temperature X-ray structures adopted the more typical cup-shaped orientation within the binding site. This supports the idea that UPGL00004 is held rigidly in the allosteric binding site of GAC, while BPTES, even when bound to the enzyme, has a significant degree of conformational flexibility. Overall, our results support a mechanism, teased out using serial room temperature crystallography, where the proper positioning of the thiadiazole rings of either compound within the hydrogen bonding network of the allosteric binding site is one of the key determinants of inhibitory potency, and that the inability to maintain the proper positioning of these rings is one reason BPTES is less potent than UPGL00004.
Broader impacts
This work highlights the importance of examining structures in conditions as close to physiological as possible. Although the molecules are still packed into a crystal in SSX, they are not frozen into shapes which might not be the most likely ones at warmer temperatures. The paper adds to the evidence that SSX has not only become a semi-routine technique, but also produces important scientific results.
Need for CHESS
The serial crystallography work at the heart of this paper can be carried out only with extensive support from the facility at which it is done, in terms of both equipment and technical support. CHESS provided such support: first, by overcoming logistical and policy hurdles to operate a beamline in June 2020, during the COVID-19 lockdown; second, by having equipment and techniques, developed in collaboration with MiTeGen, Inc., for SSX in place; third, by extensive involvement of CHESS staff with planning, collecting data, and processing the results.
Funding
The research was supported by National Institutes of Health grants GM122575, CA201402 to R.A.C.; U. Pittsburgh Medical Center Competitive Med Research Fund grant to L.A.M.; Science & Engineering Research Board (India) grant EMR/2016-002141 to N.S. Work was carried out at the Center for High Energy X-ray Sciences (CHEXS), which is supported by the National Science Foundation under award DMR-1829070, and the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award 1-P30-GM124166-01A1 from the National Institute of General Medical Sciences, National Institutes of Health, and by New York State’s Empire State Development Corporation (NYSTAR).
Reference
New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography
Shawn K Milano, Qingqiu Huang, Thuy-Tien T Nguyen, Sekar Ramachandran, Aaron Finke, Irina Kriksunov, David J Schuller, D Marian Szebenyi, Elke Arenholz, Lee A McDermott, N Sukumar, Richard A Cerione, and William P Katt
J Biol Chem 2021 298, 101535; DOI: 10.1016/j.jbc.2021.101535