What you can do with BioSAXS
What parameters can BioSAXS determine?
- radius of gyration (typically 2% accuracy)
- molecular weight (typically 10% accuracy)
- maximum intra-particle distance
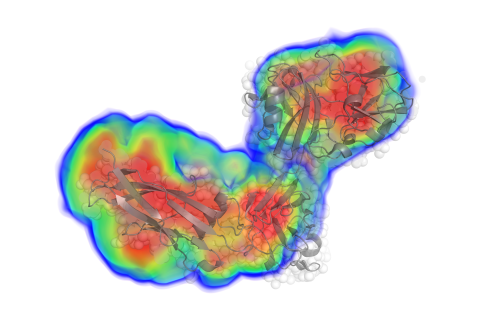
- low-resolution electron density
- degree of folding, denaturation, or disorder
- comparisons with models
- interparticle interaction potentials
What are people able to do with BioSAXS data?
- determining physiological oligomeric state
- validating proposed models of complexes
- building complexes from monomers or known fragments
- studying protein-protein interaction under different solution conditions
- modeling missing loops and domains
- refining homology models
- categorizing discrete folded and unfolded states
- finding volume fractions in mixtures
Will BioSAXS work on my samples?
In crystallography, poor crystals and overlapping spots are a frequent cause of failure. BioSAXS can have problems too. Just because it does not require crystals does not guarantee success. In crystallography, it is easier to detect bad data: you can't index or integrate the diffraction spots. With BioSAXS, however, you can process bad data with very few indications that anything is wrong. It is therefore very important to understand how to recognize bad data and to diagnose the possible causes.