What did the Scientists Discover?
Ribonucleic acid (RNA) is a biological macromolecule which performs critical roles in life. It is most widely recognized as the messenger that copies and transmits DNA’s genetic code to protein synthesizing machinery. However, RNAs biological functions extend far beyond this fundamentally important one. Many of RNA’s most remarkable tricks, e.g. its ability to self modify by cutting and pasting, result from interactions with metal ions. Given the similarity of RNA’s and DNA’s building blocks (nucleotides that contain the bases called A, C, G and U/T), these extra functions are particularly surprising because they occur in RNA and not DNA. Combining biophysical measurements that probe the conformations and electrostatics of the fundamental RNA bases, the scientists uncovered a unique energetic balance that links ion association, structure and base identity in RNA.
What are the broader impacts of this work?
These results fundamentally explain structural and functional differences between DNA and RNA that support their divergent biological roles. Through characterization of the conformations and electrostatic properties of short strands of RNA (which is negatively charged) using SAXS this work provides benchmark experimental data that enables visualization of the structures of these flexible molecules and their interactions with ion partners. In addition to providing new structures of single strands of RNA, these measurements can aid in fine tuning force fields to increase the accuracy of future simulations of RNA structure and dynamics.
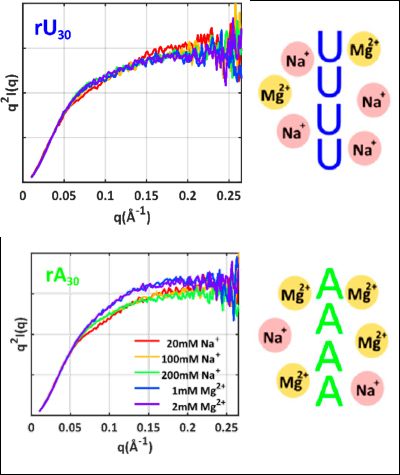
Why did this research need CHESS?
Small-angle X-ray scattering (SAXS) experiments enable the study of structural properties of these flexible and charged polymers in solution. SAXS is remarkably sensitive to the properties of the structural ensemble and its dependence on the nature of the salt ions that neutralize the negative charge of the underlying nucleic acid chain. Visualization of these ion dependent interactions provides the needed insight into the ion-base interactions that underlie RNA’s myriad of functions. SAXS experiments were conducted at CHESS beamline G1, which is ideally suited for screening multiple solution conditions.
Collaborators:
- Alex Plumridge, School of Applied and Engineering Physics Cornell University
- Kurt Andresen, Department of Physics, Gettysburg College
- Lois Pollack, School of Applied and Engineering Physics Cornell University
Publication Citation:
Plumridge, A., Andresen, K., Pollack, L., “Visualizing Disordered Single-Stranded RNA: Connecting Sequence, Structure, and Electrostatics,” J. Am. Chem. Soc.142, 109 (2020) DOI: 10.1021/jacs.9b04461
How was the work funded?
This work was supported by the National Institutes of Health [R35-GM122514]. CHESS is supported by the NSF (DMR-1332208) and the MacCHESS resource is supported by NIGMS Award No. GM-103485.