What did the Scientists Discover?
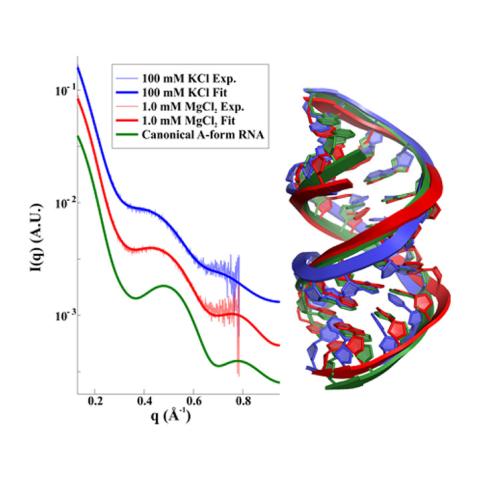
Ribonucleic acid (RNA) is a macromolecule essential in various biological roles in coding, decoding, regulation and expression of genes. Its biological functions depend critically on its structure and flexibility. To date, no consistent picture has been obtained that describes the range of conformations assumed by RNA duplexes. Here, Cornell researchers used X-ray scattering at CHESS to quantify these variations. Their results quantify the substantial and solution-dependent deviations of double-stranded (ds) RNA duplexes from the assumed canonical A-form conformation.
Why is this important? What are the boarder impacts of this work?
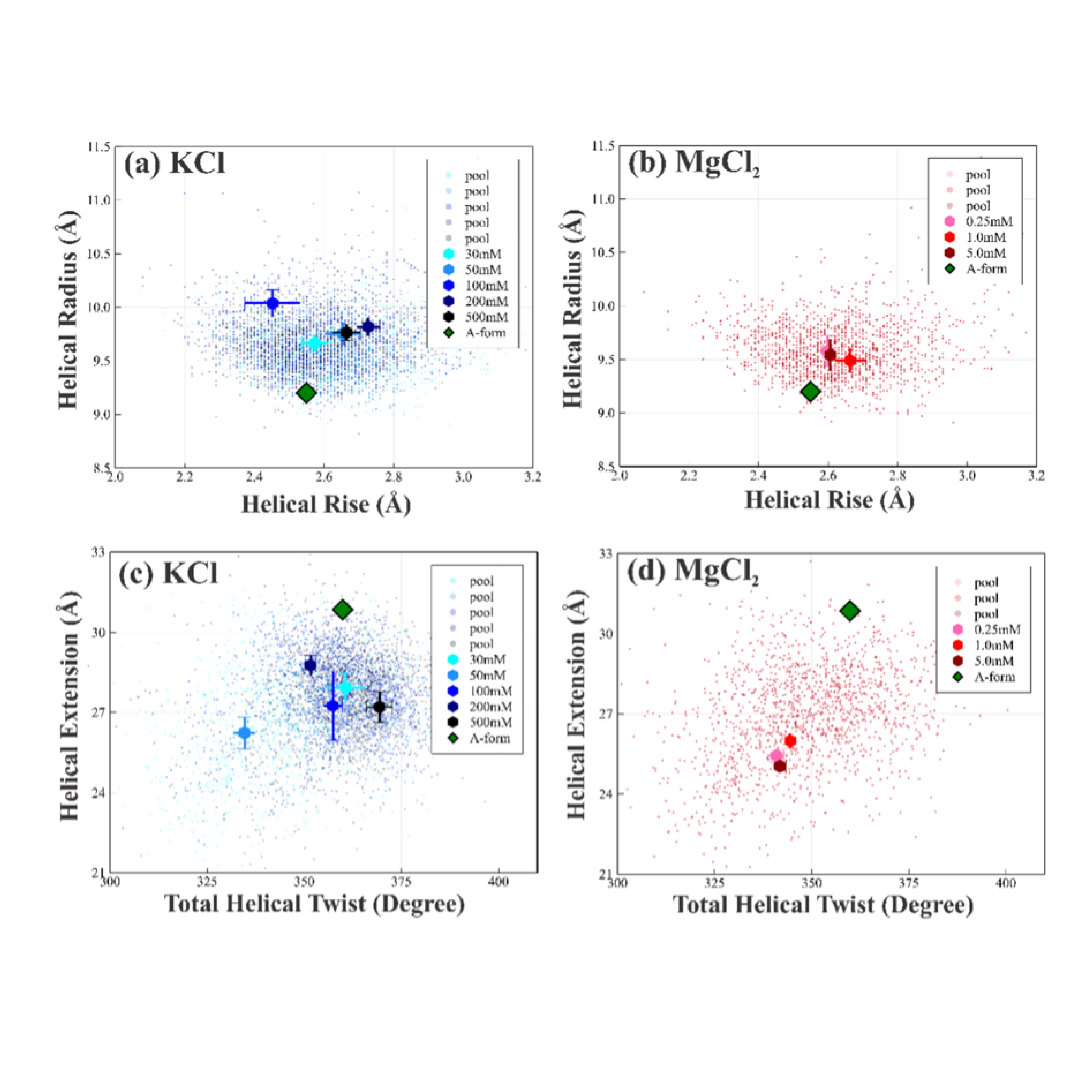
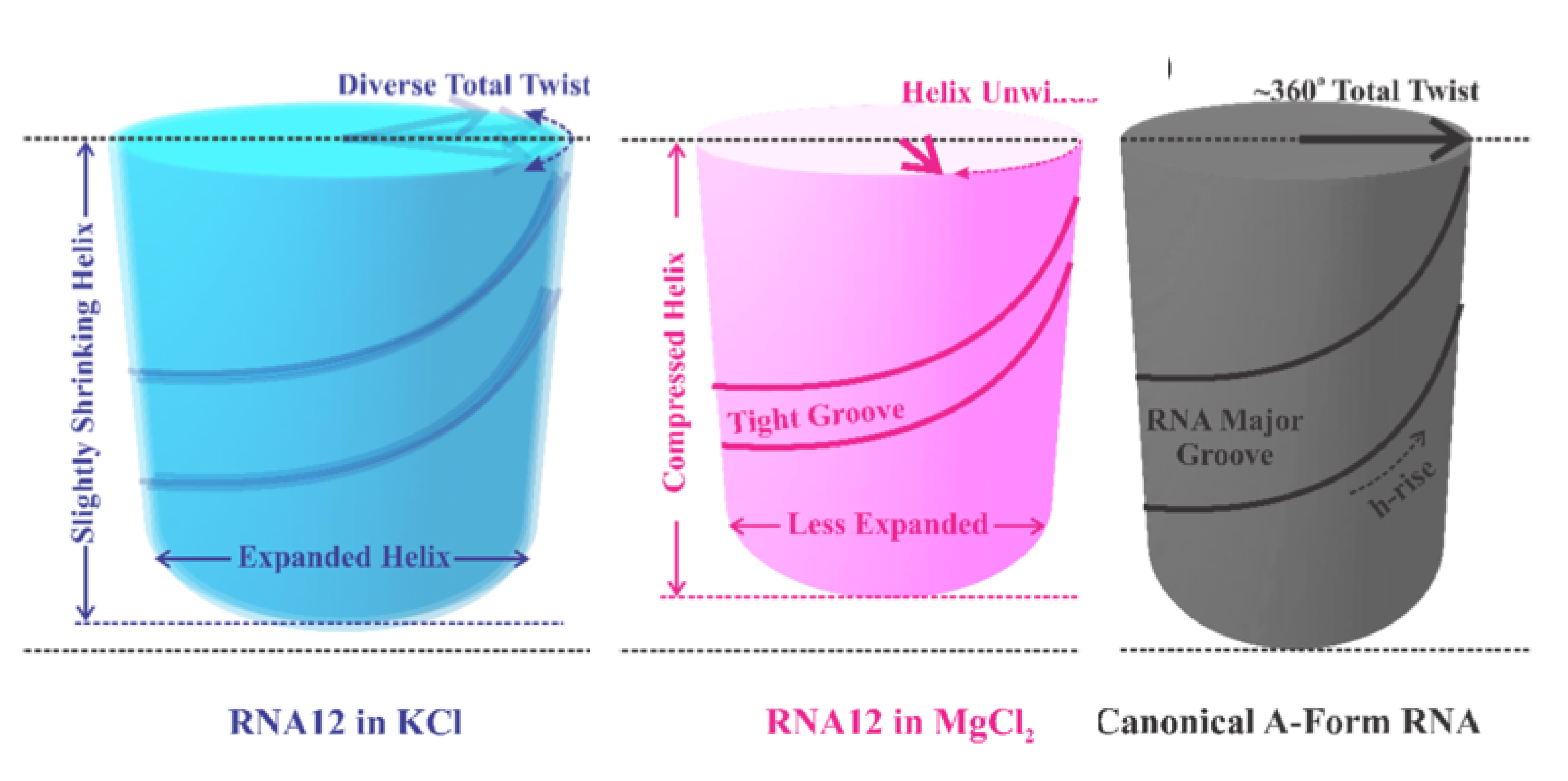
Together with DNA and proteins, RNA forms the trinity of macromolecules (large and heavy molecules) essential to all forms of life on earth. The researchers demonstrate the ability to model and extract important quantitative structural information on RNA duplexes, with an emphasis on the less explored, but information rich wider-angle X-ray scattering data. The results underscore the dynamic nature of the dsRNA duplex. These structural dynamics must be considered when accounting for any inter- or intramolecular interactions of RNA. Beyond folding, RNA duplexes serve various biological functions that rely on precisely sculpted interactions with their surfaces. All of these processes rely on understanding the structure(s) of the RNA duplex.
Why did this research need CHESS?
Wide-angle X-ray scattering (WAXS) experiments report structural variations on length scales of ∼10 Å, shorter than their counterpart small-angle X-ray scattering (SAXS). Because the helical features (e.g., width and periodicity) of dsRNA duplexes are on this length scale, WAXS is an ideal probe of their structures. SAXS and WAXS experiments were conducted at CHESS beamline G1. Scattering profiles were recorded on EigerX 1 M or PILATUS 100 K detectors (Dectris AG, Switzerland). The SAXS and WAXS profiles were matched in regions of overlapping q from 0.210 to 0.271 Å−1. Real-time SWAXS analysis was performed with the RAW26 software package. In-house MATLAB scripts were employed for off-line, detailed SWAXS data processing.
Collaborators:
- Yen-Lin Chen, School of Applied and Engineering Physics, Cornell University,
- Lois Pollack, School of Applied and Engineering Physics, Cornell University,
Publication Citation:
Y. Chen, L. Pollack, “Salt Dependence of A-Form RNA Duplexes: Structures and Implications,” J. Phys. Chem. B 2019, 123, 46, 9773-9785 https://doi.org/10.1021/acs.jpcb.9b07502
How was the work funded?
CHESS was supported by the NSF under award DMR-1332208. The work in Prof. Pollack’s research group was funded through NIH awards, R01-GM085062 and R35-GM122514. Additional support was provided by the Cornell University Center for Advanced Computing, which receives funding from Cornell University, the NSF, and members of its Partner Program.