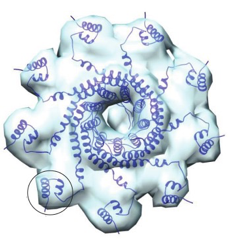
The Cingolani group (Thomas Jefferson U) has now determined the structure of TerS from the Pseudomonas phage PaP3. Phage DNA to be packaged contains multiple copies of the genome, but just one copy is needed to fill a procapsid. Terminases attempt to package this one copy by various methods; in PaP3 a termination signal is provided by the interaction of a specific sequence in the DNA (the cos sequence) with TerS.
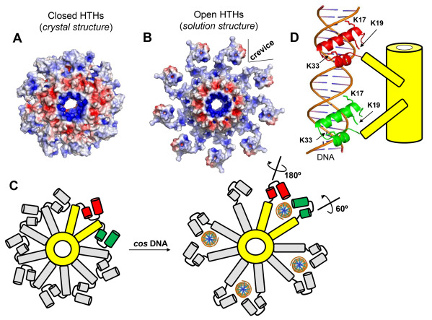
A crystal structure of PaP3 TerS reveals a nonameric ring of mixed alpha/beta composition, sitting atop a 9-stranded beta-barrel. Projecting out from the ring are spokes tipped with helix-turn-helix (HTH) DNA-binding domains. In the crystal, with no DNA present, the HTH domains are packed tightly against the inner parts of the nonamer (a "closed" form). Crystals of TerS from the related NV1 phage were also studied; their quality was not as good but the same conformation was found. BioSAXS coupled to size-exclusion chromatography, at CHESS, was then used to examine the PaP3 TerS structure, and that of the related NV1 protein, in solution. Both turned out to be ~25% larger than predicted from the crystal structure. The molecular envelope determined from SAXS data for NV1 clearly showed protuberances on the outside of the nonameric ring that did not match the crystal structure. However, by rotating the HTH domain of each monomer about an obvious hinge region, an "open" model could be built that fit the SAXS envelope well (Figure 1). The SAXS envelope for PaP3 TerS was less clear - perhaps a mixture of open and closed forms is present in solution. Biochemical studies indicated that DNA binds to the HTH domain (a suggestion that DNA might pass through the central hole in the nonamer was incompatible with this work, and with the small size of the hole), and a plausible model for DNA binding to TerS was constructed (Figure 2).
Broader impacts
Although considerable work has been done with terminases from phages that infect E. coli and Salmonella, much less has been carried out with Pseudomonas-infecting phages such as NV1 and PaP3, which are of increasing medical importance. The model determined for PaP3 is likely to apply to other phages, and will be instructive even in cases where a different mode of binding exists.
Need for CHESS
Molecular envelopes determined by small-angle X-ray scattering (SAXS) were critical to the modelling at the heart of this work. These were obtained from data collected at the CHESS BioSAXS station, ID7A, using size-exclusion chomatography (SEC) coupled with SAXS. The high flux available, from a Cornell Compact Undulator followed by a multilayer monochromator, enabled the researchers to record a good signal from the relatively dilute solution coming off the SEC column, and staff member Richard Gillilan provided excellent support to ensure that good data were obtained.
Funding
The research was supported by National Institutes of Health grants R01 GM100888, S10 OD017987, OD023479 to G.C. Work was carried out at the Center for High Energy X-ray Sciences (CHEXS), which is supported by the National Science Foundation under award DMR-1829070, and the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award 1-P30-GM124166-01A1 from the National Institute of General Medical Sciences, National Institutes of Health, and by New York State’s Empire State Development Corporation (NYSTAR).
Reference
Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation
Marzia Niazi, Tyler J Florio, Ruoyu Yang, Ravi K Lokareddy, Nicholas A Swanson, Richard E Gillilan, and Gino Cingolani
Nucleic Acids Research 2020 48, 11721-11736; DOI: 10.1093/nar/gkaa866