What did the Scientists Discover?
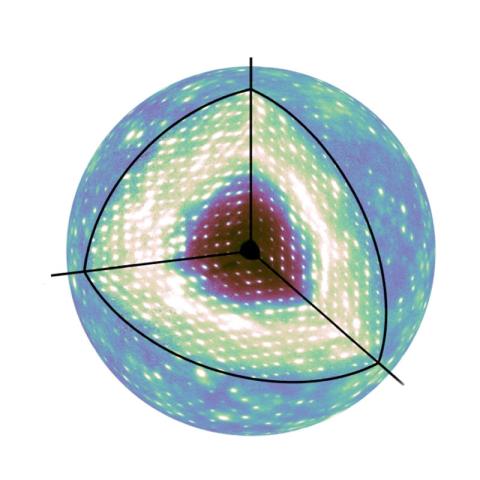
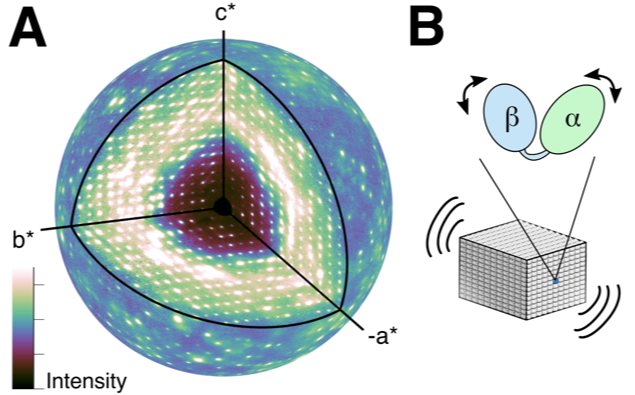
For many decades, X-ray crystallography has been the method of choice for determining the atomic-resolution structure of protein molecules arranged in a crystal lattice. X-rays diffract from the crystal, producing Bragg peaks on a detector that encode the structure. Interestingly, protein motions in the crystal give rise to a second signal, known as diffuse scattering, that appears between and underneath the Bragg peaks. However, this signal has been challenging to measure and interpret. Working at CHESS, the Ando group at Cornell succeeded in mapping the three-dimensional diffuse scattering from a protein crystal with unprecedented accuracy (Fig. 1). Using this high-quality map, they were able to show that lattice vibrations were responsible for most of the diffuse pattern, including the striking “halo” features near the Bragg peaks (Fig. 2a). Once these motions were accounted for, they showed that internal breathing motions of the protein contribute in a subtle but important way (Fig. 2b).
Why is this important? What are the broader impacts of this work?
It is increasingly recognized in the field of structural biology that information on protein dynamics is needed to understand function, but few techniques are sensitive to the dynamics of interest. Diffuse scattering has been proposed to fill this critical gap, since it is often observed during the course of conventional data collection and could be used to "animate" conventional crystal structures. However, diffuse scattering is routinely discarded at the first step in data processing, because it has proven exceedingly difficult to interpret. This work provides the first convincing demonstration of protein diffuse scattering data collection and analysis, opening the door to future applications in structural biology.
Why did this research need CHESS?
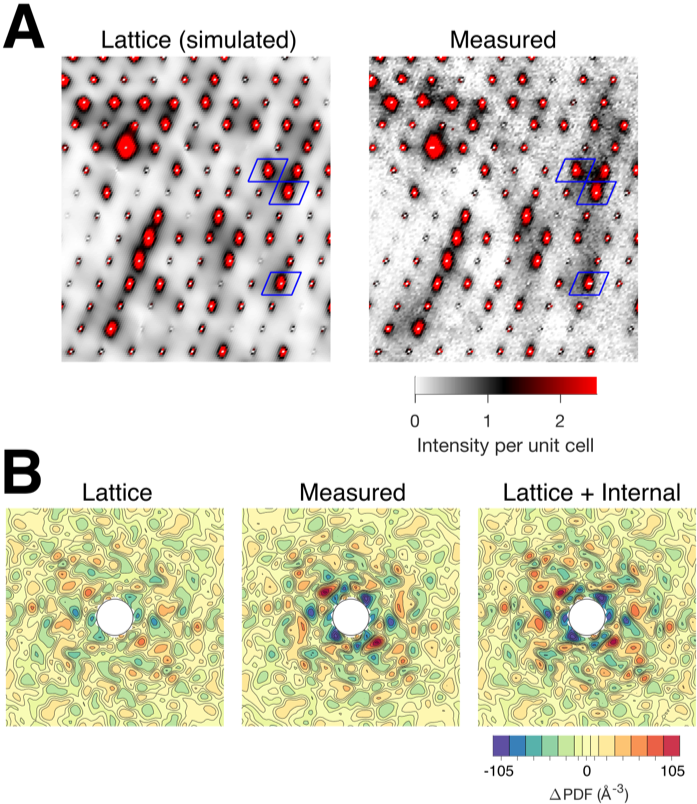
Many of the advances that made this work possible have occurred at CHESS, which has a long history in the field of protein diffuse scattering. Since the 1990s, Sol Gruner and his group have pioneered the use of CCDs and later, pixel-array detectors for diffuse scattering. Additionally, CHESS's unique "empty hutch" culture of experimentation makes it the ideal environment for performing non-standard experiments and developing new methods. The diffuse scattering data in this paper were collected at room temperature on the MacCHESS F1 beamline using the Pilatus 6M pixel-array detector. It was by taking full advantage of the F1 hutch and the detector's performance that the authors were able to improve data quality to the point where realistic models could be fit.
Collaborators:
- Steve P. Meisburger, Department of Chemistry, Princeton University, Princeton, NJ, 08544, USA & Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, 14850, USA
- David A. Case, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ, 08854, USA
- Nozomi Ando, Department of Chemistry, Princeton University, Princeton, NJ, 08544, USA & Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, 14850, USA
Publication Citation:
Meisburger, S.P., Case, D.A. & Ando, N., “Diffuse X-ray scattering from correlated motions in a protein crystal.” Nat Commun 11, 1271 (2020). https://doi.org/10.1038/s41467-020-14933-6
How was the work funded?
CHESS is supported by NSF Grant DMR-1332208, and the MacCHESS facility is supported by NIH/NIGMS Grant GM-103485. The authors were supported by NIH Grants GM117757 (to S.P.M.), GM100008 (to N.A.), GM124847 (to N.A.), and GM122086 (to D.A.C.) and by start-up funds from Princeton University and Cornell University (to N.A.).