PyMOL is a popular molecular visualization software used by scientists and researchers to visualize and analyze molecular structures. It is designed to work with a variety of file formats, including PDB (Protein Data Bank) files, and can display a range of molecular properties, such as protein linkage, electrostatic interactions and hydrogen bonding.
The Cornell High Energy Synchrotron Source (CHESS) facility located at Cornell University provides intense beams of X-rays for use in scientific imaging research, such as the study of atomic and molecular structures of materials and biological samples. While other synchrotron laboratories are traditionally located at national labs, Cornell is the only U.S. university still operating a large accelerator complex – CHESS.
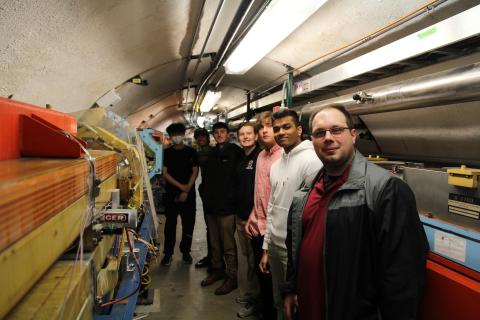
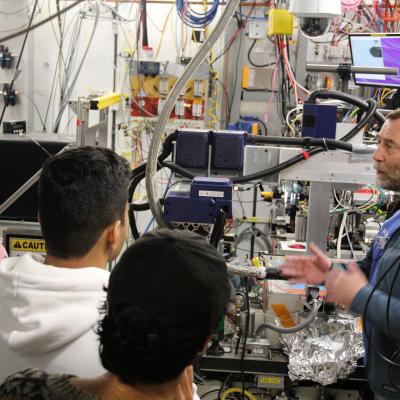
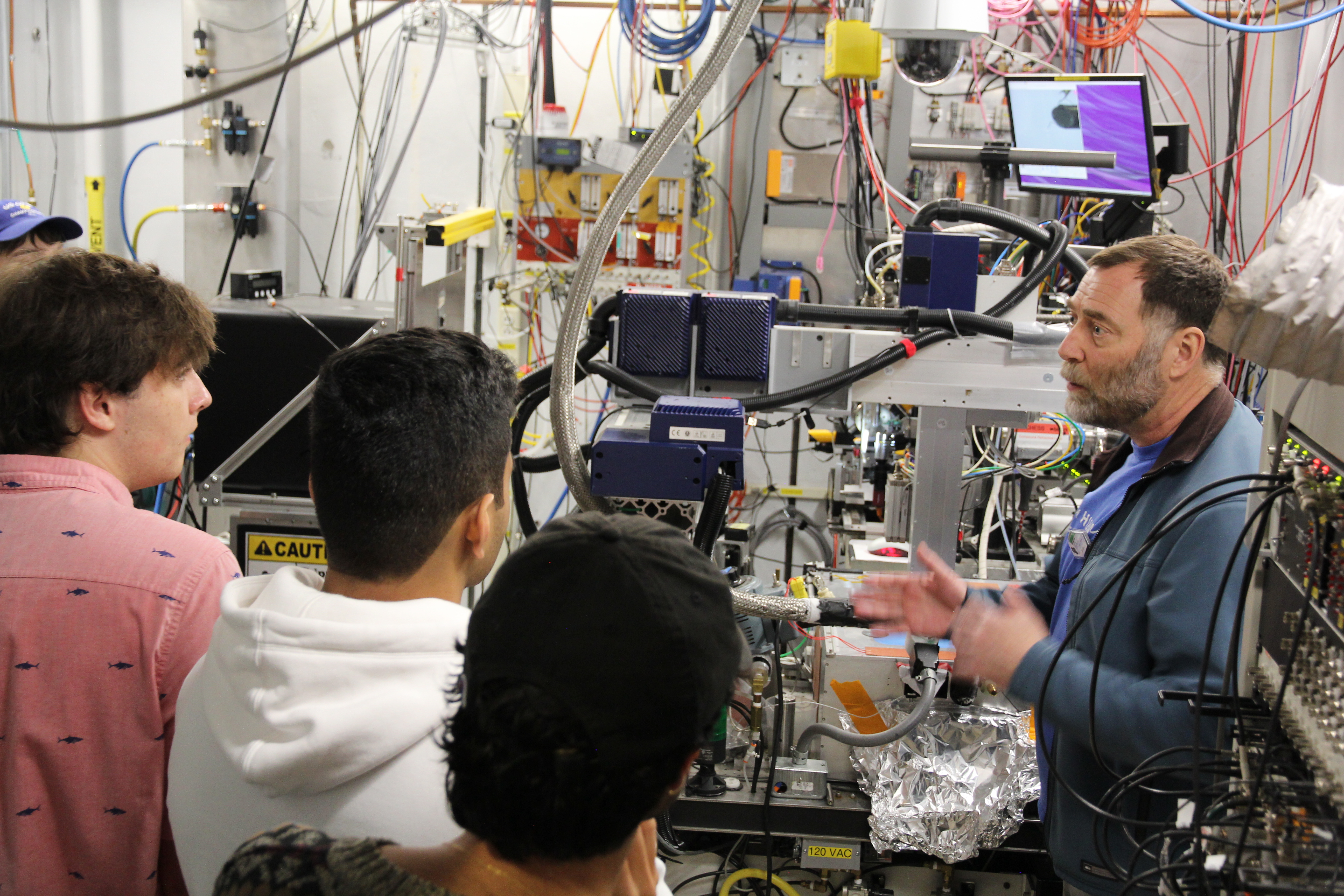
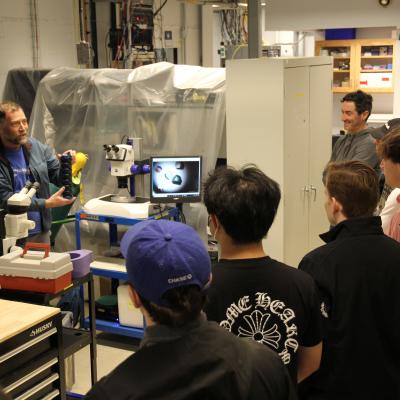
Cornell students studying various fields of biological sciences came to CHESS to see this first-hand with their instructor, Kevin Siegenthaler, to learn more about the facility’s relevance to PyMOL. They went on an in depth tour led by Rick Ryan and David Schuller down into the underground storage ring, the sector 7 biological beamlines, and the Wilson Laboratory scientific offices. This rare opportunity allowed the students to see the facility infrastructure that is essential in generating a 3D molecular model and to appreciate the years of data collection and analysis that this process requires.
The connection between PyMOL and CHESS lies in their complementary roles in the field of structural biology. PyMOL is used to visualize the shape of molecular structures, while CHESS provides the X-ray diffraction data used to determine these structures.
When scientists at CHESS study the structure of a molecule, they collect X-ray diffraction data that can be used to determine the positions of individual atoms within the molecule. This data is then analyzed using computational methods to generate a 3D model of the molecule. PyMOL is often used to visualize these models, allowing researchers to explore the structure in greater detail and gain insights into its function.
In addition to visualizing structures determined by X-ray crystallography, PyMOL can also be used to generate models of molecular structures using other techniques, such as NMR (Nuclear Magnetic Resonance) spectroscopy. These models can then be refined using X-ray crystallography data obtained from CHESS.
By combining experimental data from synchrotron sources like CHESS with computational tools like PyMOL, scientists can gain a more complete understanding of the molecular structures that underlie biological processes. CHESS has contributed over 2,000 protein structures to the Protein Data Bank, with great promise to continually enrich this open-source community.