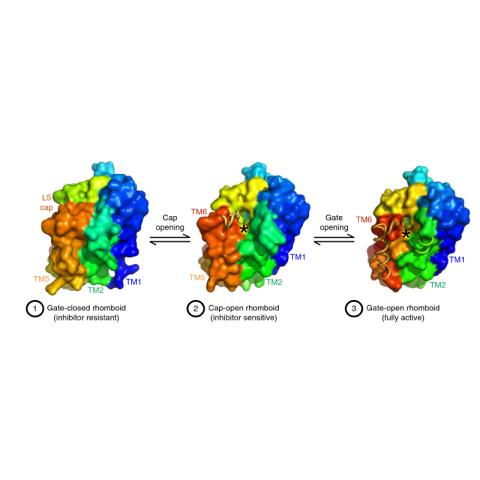
What did the Scientists Discover?
Protein cleavage inside the cell membrane triggers various signaling pathways, but the mechanism of catalysis is poorly understood. Here, using crystallography capabilities at CHESS, the researchers solved ten structures of the Escherichia coli rhomboid protease, embedded in a bicelle mimic of a cell membrane and reacting with either an inhibitor or an actual peptide substrate. The time-resolved steps observed encompass the entire proteolytic reaction catalyzed by the enzyme. Extensive gate opening accompanied substrate, but not inhibitor, binding, revealing that substrates and inhibitors take different paths to the active site. Catalysis unexpectedly commenced with, and was guided through subsequent catalytic steps by, motions of an extracellular loop, with local contributions from active site residues. The authors even captured the elusive tetrahedral intermediate that is uncleaved but covalently attached to the catalytic serine, about which the substrate was forced to bend dramatically. This unexpectedly stable intermediate indicates that rhomboid catalysis uses an unprecedented reaction coordinate that may involve mechanically stressing the peptide bond, and could be selectively targeted by inhibitors.
Why is this important?
Four large and distinct superfamilies of intramembrane proteases evolved to regulate diverse signaling pathways from bacteria to humans. These unusual enzymes have been implicated in diseases ranging from Alzheimer’s disease to malaria, type II diabetes, cancer, Parkinson’s disease, cholera and tuberculosis. Despite their prominent pathophysiological roles, the steps underlying catalysis have never been observed directly for any intramembrane protease; knowledge of these will be crucial to eventually develop cures for these diseases. Time-resolved crystallography can visualize the discrete actions taken by enzymes during catalysis and thereby directly map out the reaction coordinate.
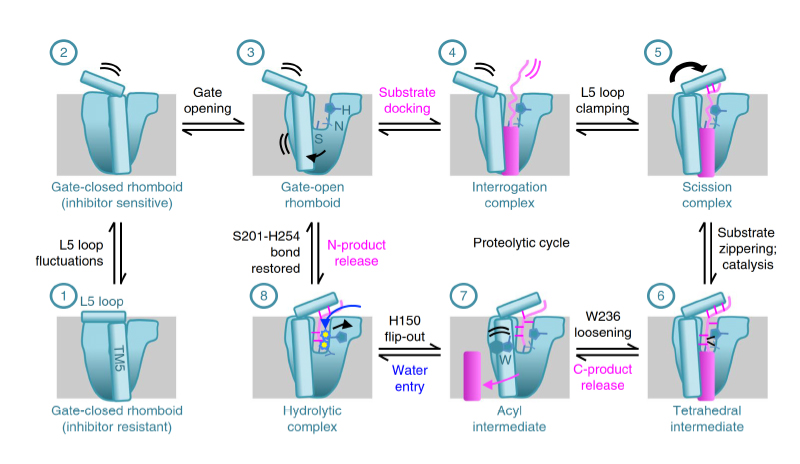
Why did this research need CHESS?
All X-ray diffraction data were collected using the F1 station at CHESS dedicated to macromolecular crystallography and operated by MacCHESS. The FlexX beamline which is part of the NSF-funded Center for High Energy X-ray Sciences (CHEXS) at CHESS and as well as the NIH/NYSTAR funded MacCHESS will allow similar studies in the future.
Collaborators:
- Sangwoo Cho, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore./li>
- Rosanna P. Baker, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore.
- Ming Ji, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore.
- Siniša Urban, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore.
Publication Citation:
Cho, S., Baker, R.P., Ji, M. et al., “Ten catalytic snapshots of rhomboid intramembrane proteolysis from gate opening to peptide release.” Nat Struct Mol Biol, vol. 26, pp. 910-918, Oct. 2019. https://doi.org/10.1038/s41594-019-0296-9
How was the work funded?
This work was supported by a National Institutes of Health grant (no. R01AI066025 to S.U.). CHESS was supported by NSF grant DMR-0936384 and National Institutes of Health grant GM-103485).